edtools Demo
edtools is a python package for automated processing of a large number of 3D electron diffraction (3D ED) datasets. It can be downloaded from https://doi.org/10.5281/zenodo.6952810.
For runing edtools, XDS package for reduction of 3D ED datasets is required. XDS package is available at https://xds.mr.mpg.de/html_doc/downloading.html.
A typical cycle of using edtools for processing batch 3D ED datasets goes through the following steps:
edtools.autoindexedtools.extract_xds_infoedtools.find_celledtools.update_xdsedtools.make_xscaleedtools.cluster
Here we demonstrate the processing of batch 3D ED datasets for phase analysis and structure determination using edtools. The datasets for the demo can be downloaded from https://zenodo.org/record/6533426#.YnoQ7_hBxaQ.
The datasets were collected on a zeolite mixture sample using serial rotation electron diffraction (SerialRED) data collection technique implemented in the program Instamatic (available at https://doi.org/10.5281/zenodo.5175957), which runs on a JEOL JEM-2100-LaB6 at 200 kV equipped with a 512 x 512 Timepix hybrid pixel detector (55 x 55 µm pixel size, QTPX-262k, Amsterdam Scientific Instruments).
The zeolite mixture sample contains phases IWV,RTH, and *CTH. The information of these three phases can be found from the structure database of zeolites (https://europe.iza-structure.org/IZA-SC/ftc_table.php).
This demo takes around 5-10 min to run on a normal desktop computer with all the required packages installed properly beforehand.
Indexing
Automatically index the 3D ED datasets by running XDS in all subfolders (SMV) that contains file XDS.INP, which is automatically generated during data collection using Instamatic.
[1]:
!edtools.autoindex
!!! ERROR !!! WRONG TYPE OF INPUT FILE SPECIFIED
!!! ERROR !!! WRONG TYPE OF INPUT FILE SPECIFIED
16 files named XDS.INP (subdir: None) found.
0: C:\demo\edtools_demo_data\stagepos_0067\crystal_0001\SMV # Mon Aug 1 21:00:56 2022
Spgr 5 - Cell 26.93 14.05 5.36 90.00 90.89 90.00 - Vol 2027.80
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
---------------------------------------------------------------------------------
0 4.35 0.80 583 324 15.0 4.59 13.7 99.0 7.47 6.72
- 0.85 0.80 54 42 12.5 1.96 26.8 91.9
1: C:\demo\edtools_demo_data\stagepos_0164\crystal_0000\SMV # Mon Aug 1 21:00:58 2022
Spgr 1 - Cell 9.49 9.90 12.47 66.56 89.45 86.35 - Vol 1072.59
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
1 6.39 0.80 229 209 4.8 24.01 22.3 96.3 50.00 4.74
- 0.91 0.85 31 29 4.5 12.16 21.3 0.0
3: C:\demo\edtools_demo_data\stagepos_0299\crystal_0001\SMV # Mon Aug 1 21:01:00 2022
Spgr 1 - Cell 4.83 14.83 16.03 115.66 89.61 94.16 - Vol 1031.87
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
3 2.05 0.80 400 312 7.5 2.44 20.6 95.2 4.24 6.11
- 0.84 0.80 27 26 4.7 1.79 11.2 0.0
4: C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV # Mon Aug 1 21:01:02 2022
Spgr 5 - Cell 13.69 25.42 14.90 90.00 115.84 90.00 - Vol 4666.87
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
4 11.09 0.79 3744 2147 42.8 3.44 13.1 99.6 13.90 8.10
- 0.97 0.90 623 336 47.9 1.32 68.9 84.7
5: C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV # Mon Aug 1 21:01:03 2022
Spgr 5 - Cell 25.67 13.50 17.73 90.00 132.44 90.00 - Vol 4534.43
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
5 6.86 0.80 2161 1081 21.8 4.23 10.5 99.9 33.30 8.74
- 0.97 0.90 342 159 23.3 0.83 130.3 69.6
6: C:\demo\edtools_demo_data\stagepos_0368\crystal_0001\SMV # Mon Aug 1 21:01:05 2022
Spgr 1 - Cell 10.17 10.36 12.16 93.71 113.40 98.01 - Vol 1154.16
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
6 10.17 0.80 611 443 9.4 3.17 14.9 97.5 5.07 4.64
- 0.85 0.80 56 53 7.0 1.96 73.1 6.4
7: C:\demo\edtools_demo_data\stagepos_0538\crystal_0000\SMV # Mon Aug 1 21:01:06 2022
Spgr 1 - Cell 10.55 10.52 11.81 80.39 66.60 75.74 - Vol 1162.33
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
7 5.10 0.80 443 330 7.0 3.80 10.7 99.4 8.61 5.62
- 0.85 0.80 38 36 4.8 1.80 76.5 0.0
8: C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV # Mon Aug 1 21:01:07 2022
Spgr 1 - Cell 13.82 14.32 16.18 86.20 111.75 116.39 - Vol 2645.41
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
8 6.37 0.80 1460 989 9.1 2.88 16.3 97.8 5.24 7.62
- 0.85 0.80 166 125 7.3 1.36 62.8 56.0
9: C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV # Mon Aug 1 21:01:08 2022
Spgr 5 - Cell 15.06 26.22 15.41 90.00 118.30 90.00 - Vol 5357.50
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
9 13.11 0.79 2063 1319 22.1 3.46 10.5 99.6 12.09 7.58
- 0.89 0.83 326 223 24.5 1.01 53.6 83.4
10: C:\demo\edtools_demo_data\stagepos_0905\crystal_0000\SMV # Mon Aug 1 21:01:10 2022
Spgr 3 - Cell 13.91 5.07 14.97 90.00 117.96 90.00 - Vol 932.53
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
10 12.33 0.80 479 300 13.8 3.68 13.4 99.6 16.07 9.46
- 1.20 1.07 58 35 14.6 4.72 24.9 88.5
11: C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV # Mon Aug 1 21:01:11 2022
Spgr 1 - Cell 13.71 14.57 15.77 83.07 68.29 62.34 - Vol 2587.36
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
11 11.49 0.80 1596 1144 10.7 3.30 12.4 98.5 7.24 7.18
- 0.85 0.80 124 121 7.0 0.94 22.6 83.4
12: C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV # Mon Aug 1 21:01:13 2022
Spgr 1 - Cell 14.56 15.00 15.27 97.22 105.97 120.36 - Vol 2621.77
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
12 7.54 0.80 1746 1222 11.3 4.00 13.3 98.6 8.77 5.85
- 0.85 0.80 164 146 8.4 1.48 36.5 86.5
13: C:\demo\edtools_demo_data\stagepos_1014\crystal_0000\SMV # Mon Aug 1 21:01:14 2022
Spgr 1 - Cell 5.30 14.56 15.04 112.06 93.44 86.65 - Vol 1072.87
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
13 5.01 0.81 447 328 7.5 4.11 10.9 98.5 6.65 6.67
- 0.85 0.80 51 44 6.3 2.11 18.7 92.7
15: C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV # Mon Aug 1 21:01:17 2022
Spgr 1 - Cell 13.64 15.02 25.09 93.07 91.13 114.33 - Vol 4672.25
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
15 6.60 0.80 3124 2149 11.3 3.54 8.4 99.5 12.64 6.94
- 0.85 0.80 346 280 9.2 1.24 56.2 84.9
Extract cell
Extract the determined unit cell parameters from the output files (CORRECT.LP) of XDS
[2]:
!edtools.extract_xds_info
14 files named CORRECT.LP (subdir: None) found.
1: C:\demo\edtools_demo_data\stagepos_0067\crystal_0001\SMV # Mon Aug 1 21:00:56 2022
Spgr 5 - Cell 26.93 14.05 5.36 90.00 90.89 90.00 - Vol 2027.80
2: C:\demo\edtools_demo_data\stagepos_0164\crystal_0000\SMV # Mon Aug 1 21:00:58 2022
Spgr 1 - Cell 9.49 9.90 12.47 66.56 89.45 86.35 - Vol 1072.59
3: C:\demo\edtools_demo_data\stagepos_0299\crystal_0001\SMV # Mon Aug 1 21:01:00 2022
Spgr 1 - Cell 4.83 14.83 16.03 115.66 89.61 94.16 - Vol 1031.87
4: C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV # Mon Aug 1 21:01:02 2022
Spgr 5 - Cell 13.69 25.42 14.90 90.00 115.84 90.00 - Vol 4666.87
5: C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV # Mon Aug 1 21:01:03 2022
Spgr 5 - Cell 25.67 13.50 17.73 90.00 132.44 90.00 - Vol 4534.43
6: C:\demo\edtools_demo_data\stagepos_0368\crystal_0001\SMV # Mon Aug 1 21:01:05 2022
Spgr 1 - Cell 10.17 10.36 12.16 93.71 113.40 98.01 - Vol 1154.16
7: C:\demo\edtools_demo_data\stagepos_0538\crystal_0000\SMV # Mon Aug 1 21:01:06 2022
Spgr 1 - Cell 10.55 10.52 11.81 80.39 66.60 75.74 - Vol 1162.33
8: C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV # Mon Aug 1 21:01:07 2022
Spgr 1 - Cell 13.82 14.32 16.18 86.20 111.75 116.39 - Vol 2645.41
9: C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV # Mon Aug 1 21:01:08 2022
Spgr 5 - Cell 15.06 26.22 15.41 90.00 118.30 90.00 - Vol 5357.50
10: C:\demo\edtools_demo_data\stagepos_0905\crystal_0000\SMV # Mon Aug 1 21:01:10 2022
Spgr 3 - Cell 13.91 5.07 14.97 90.00 117.96 90.00 - Vol 932.53
11: C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV # Mon Aug 1 21:01:11 2022
Spgr 1 - Cell 13.71 14.57 15.77 83.07 68.29 62.34 - Vol 2587.36
12: C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV # Mon Aug 1 21:01:13 2022
Spgr 1 - Cell 14.56 15.00 15.27 97.22 105.97 120.36 - Vol 2621.77
13: C:\demo\edtools_demo_data\stagepos_1014\crystal_0000\SMV # Mon Aug 1 21:01:14 2022
Spgr 1 - Cell 5.30 14.56 15.04 112.06 93.44 86.65 - Vol 1072.87
14: C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV # Mon Aug 1 21:01:17 2022
Spgr 1 - Cell 13.64 15.02 25.09 93.07 91.13 114.33 - Vol 4672.25
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
---------------------------------------------------------------------------------
1 4.35 0.80 583 324 15.0 4.59 13.7 99.0 7.47 6.72 # C:\demo\edtools_demo_data\stagepos_0067\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 54 42 12.5 1.96 26.8 91.9
2 6.39 0.80 229 209 4.8 24.01 22.3 96.3 50.00 4.74 # C:\demo\edtools_demo_data\stagepos_0164\crystal_0000\SMV\CORRECT.LP
- 0.91 0.85 31 29 4.5 12.16 21.3 0.0
3 2.05 0.80 400 312 7.5 2.44 20.6 95.2 4.24 6.11 # C:\demo\edtools_demo_data\stagepos_0299\crystal_0001\SMV\CORRECT.LP
- 0.84 0.80 27 26 4.7 1.79 11.2 0.0
4 11.09 0.79 3744 2147 42.8 3.44 13.1 99.6 13.90 8.10 # C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV\CORRECT.LP
- 0.97 0.90 623 336 47.9 1.32 68.9 84.7
5 6.86 0.80 2161 1081 21.8 4.23 10.5 99.9 33.30 8.74 # C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV\CORRECT.LP
- 0.97 0.90 342 159 23.3 0.83 130.3 69.6
6 10.17 0.80 611 443 9.4 3.17 14.9 97.5 5.07 4.64 # C:\demo\edtools_demo_data\stagepos_0368\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 56 53 7.0 1.96 73.1 6.4
7 5.10 0.80 443 330 7.0 3.80 10.7 99.4 8.61 5.62 # C:\demo\edtools_demo_data\stagepos_0538\crystal_0000\SMV\CORRECT.LP
- 0.85 0.80 38 36 4.8 1.80 76.5 0.0
8 6.37 0.80 1460 989 9.1 2.88 16.3 97.8 5.24 7.62 # C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 166 125 7.3 1.36 62.8 56.0
9 13.11 0.79 2063 1319 22.1 3.46 10.5 99.6 12.09 7.58 # C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV\CORRECT.LP
- 0.89 0.83 326 223 24.5 1.01 53.6 83.4
10 12.33 0.80 479 300 13.8 3.68 13.4 99.6 16.07 9.46 # C:\demo\edtools_demo_data\stagepos_0905\crystal_0000\SMV\CORRECT.LP
- 1.20 1.07 58 35 14.6 4.72 24.9 88.5
11 11.49 0.80 1596 1144 10.7 3.30 12.4 98.5 7.24 7.18 # C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 124 121 7.0 0.94 22.6 83.4
12 7.54 0.80 1746 1222 11.3 4.00 13.3 98.6 8.77 5.85 # C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV\CORRECT.LP
- 0.85 0.80 164 146 8.4 1.48 36.5 86.5
13 5.01 0.81 447 328 7.5 4.11 10.9 98.5 6.65 6.67 # C:\demo\edtools_demo_data\stagepos_1014\crystal_0000\SMV\CORRECT.LP
- 0.85 0.80 51 44 6.3 2.11 18.7 92.7
14 6.60 0.80 3124 2149 11.3 3.54 8.4 99.5 12.64 6.94 # C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 346 280 9.2 1.24 56.2 84.9
Wrote 14 cells to file cells.xlsx
Wrote 14 cells to file cells.yaml
Wrote 8 entries to file filelist.txt (completeness > 10.0%, CC(1/2) > 90.0%)
Most likely lattice types:
1 Lattice type `aP` (spgr: 1) was found 9 times (score: 10056)
2 Lattice type `mC` (spgr: 5) was found 4 times (score: 8551)
3 Lattice type `mP` (spgr: 3) was found 1 times (score: 479)
** the score corresponds to the total number of indexed reflections.
Unit-cell-based clustering for phase analysis
[ ]:
!edtools.find_cell cells.yaml -s --cluster --metric lcv
[3]:
from IPython.display import Image
Image('find_cell_step3.png', embed=True)
[3]:
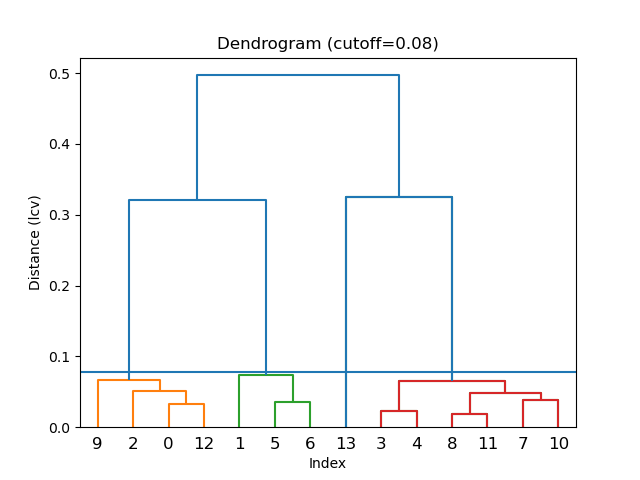
Console Output
Linkage method = average
Cutoff distance = 0.078
Distance metric = lcv
----------------------------------------
Cluster #1 (4 items)
1 [ 5.47 14.07 15.30 63.22 87.59 88.58] Vol.: 1050.9
3 [ 5.33 14.99 16.06 64.44 89.16 82.51] Vol.: 1144.9
10 [ 5.05 14.37 14.53 62.13 88.52 89.11] Vol.: 932.0
13 [ 5.30 14.89 15.18 66.79 86.51 86.59] Vol.: 1098.1
---
Mean: [ 5.29 14.58 15.27 64.15 87.95 86.70] Vol.: 1056.5
Min: [ 5.05 14.07 14.53 62.13 86.51 82.51] Vol.: 932.0
Max: [ 5.47 14.99 16.06 66.79 89.16 89.11] Vol.: 1144.9
Cluster #2 (3 items)
2 [ 9.52 9.98 12.85 65.60 87.80 85.43] Vol.: 1107.8
6 [ 10.21 10.36 12.08 85.86 67.02 81.83] Vol.: 1165.3
7 [ 10.55 10.75 11.75 80.34 66.42 75.73] Vol.: 1179.4
---
Mean: [ 10.09 10.36 12.23 77.27 73.75 81.00] Vol.: 1150.9
Min: [ 9.52 9.98 11.75 65.60 66.42 75.73] Vol.: 1107.8
Max: [ 10.55 10.75 12.85 85.86 87.80 85.43] Vol.: 1179.4
Cluster #3 (6 items)
4 [ 14.04 14.39 14.72 76.68 62.79 61.86] Vol.: 2331.3
5 [ 13.50 14.38 14.63 75.73 64.60 63.07] Vol.: 2283.0
8 [ 13.89 14.29 17.00 72.43 63.61 63.57] Vol.: 2684.8
9 [ 14.81 15.07 15.52 62.45 74.78 62.16] Vol.: 2711.1
11 [ 13.73 14.56 16.03 84.26 68.05 62.57] Vol.: 2629.5
12 [ 14.43 14.90 15.40 81.24 74.01 61.15] Vol.: 2787.8
---
Mean: [ 14.07 14.60 15.55 75.46 67.97 62.40] Vol.: 2571.3
Min: [ 13.50 14.29 14.63 62.45 62.79 61.15] Vol.: 2283.0
Max: [ 14.81 15.07 17.00 84.26 74.78 63.57] Vol.: 2787.8
Wrote cluster 1 to file `cells_cluster_1_4-items.yaml`
Wrote cluster 2 to file `cells_cluster_2_3-items.yaml`
Wrote cluster 3 to file `cells_cluster_3_6-items.yaml`
The three resulted clusters 1, 2, 3 correspond to phases *CTH, RTH, and IWV, respectively.
With the averaged primitive unit cell parameters of each cluster, one can use the online tool http://cci.lbl.gov/cctbx/lattice_symmetry.html to find unit cell with higher symmetry with a pre-set tolerance.
We take cluster 3 (phase IWV) as an example. The averaged unit cell parameters are: 14.07, 14.6, 15.55, 75.46, 67.97, 62.40
The unit cell parameters with a higher symmetry (space group: Fmmm (69)) are: 14.07, 25.8828, 28.9294, 90, 90, 90
The same operation can be done for all the other clusters.
Update the XDS.INP files
This step used edtools.update_xds to update the XDS input files with the determined unit cell parameters and space group.
[4]:
!edtools.update_xds -c 14.07 25.8828 28.9294 90 90 90 -s 69
16 files named XDS.INP (subdir: None) found.
C:\demo\edtools_demo_data\stagepos_0067\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0164\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0290\crystal_0002\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0299\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0368\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0538\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0905\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_1014\crystal_0000\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_1261\crystal_0001\SMV\XDS.INP
C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV\XDS.INP
Updated 16 files
Refine phases
Rerun autoindex, extract_xds_info and find_cell for the desired phases to be successfully indexed by XDS. All the other phases are hopefully excluded in that a phase with different enough unit cell will not be indexed successfully. There are however cases when different phases have similar unit cells, which cannot be told apart during this step.
[5]:
!edtools.autoindex
!!! ERROR !!! WRONG TYPE OF INPUT FILE SPECIFIED
!!! ERROR !!! WRONG TYPE OF INPUT FILE SPECIFIED
16 files named XDS.INP (subdir: None) found.
4: C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV # Mon Aug 1 21:03:22 2022
Spgr 69 - Cell 13.88 25.44 27.26 90.00 90.00 90.00 - Vol 9625.70
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
4 9.30 0.80 3938 1852 69.0 3.24 20.2 99.4 11.45 8.21
- 0.91 0.85 614 290 74.4 0.86 109.5 81.7
5: C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV # Mon Aug 1 21:03:24 2022
Spgr 69 - Cell 13.52 24.94 27.07 90.00 90.00 90.00 - Vol 9127.70
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
5 10.88 0.80 2203 1029 40.4 3.84 11.7 99.9 27.38 9.78
- 1.07 0.98 299 135 41.8 1.04 107.2 76.5
8: C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV # Mon Aug 1 21:03:28 2022
Spgr 69 - Cell 14.01 25.97 29.04 90.00 90.00 90.00 - Vol 10565.90
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
8 7.14 0.80 1466 781 26.2 2.61 18.2 97.2 4.73 7.15
- 0.84 0.80 142 92 19.9 0.98 62.3 52.7
9: C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV # Mon Aug 1 21:03:30 2022
Spgr 69 - Cell 15.10 26.02 26.72 90.00 90.00 90.00 - Vol 10498.34
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
9 7.24 0.80 1994 1126 38.5 3.27 11.9 99.5 12.91 8.08
- 0.98 0.90 322 166 41.2 1.27 70.2 89.0
10: C:\demo\edtools_demo_data\stagepos_0905\crystal_0000\SMV -> Error in IDXREF: RETURN CODE IS IER= 0
11: C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV # Mon Aug 1 21:03:32 2022
Spgr 69 - Cell 13.83 25.80 28.73 90.00 90.00 90.00 - Vol 10251.27
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
11 7.08 0.80 1591 808 28.2 2.88 17.1 98.1 6.24 7.63
- 0.90 0.85 254 128 30.4 1.17 42.6 95.4
12: C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV # Mon Aug 1 21:03:34 2022
Spgr 69 - Cell 14.39 25.16 28.10 90.00 90.00 90.00 - Vol 10173.67
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
12 5.12 0.80 1669 851 30.2 3.75 16.8 98.0 6.26 5.76
- 0.85 0.80 153 109 25.3 1.34 46.1 68.5
15: C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV # Mon Aug 1 21:03:39 2022
Spgr 69 - Cell 13.54 25.23 27.30 90.00 90.00 90.00 - Vol 9326.07
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
15 5.97 0.80 1620 563 21.7 6.15 8.4 99.8 11.79 7.17
- 0.85 0.80 187 78 19.1 2.24 45.1 97.9
[6]:
!edtools.extract_xds_info
7 files named CORRECT.LP (subdir: None) found.
1: C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV # Mon Aug 1 21:03:22 2022
Spgr 69 - Cell 13.88 25.44 27.26 90.00 90.00 90.00 - Vol 9625.70
2: C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV # Mon Aug 1 21:03:24 2022
Spgr 69 - Cell 13.52 24.94 27.07 90.00 90.00 90.00 - Vol 9127.70
3: C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV # Mon Aug 1 21:03:28 2022
Spgr 69 - Cell 14.01 25.97 29.04 90.00 90.00 90.00 - Vol 10565.90
4: C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV # Mon Aug 1 21:03:30 2022
Spgr 69 - Cell 15.10 26.02 26.72 90.00 90.00 90.00 - Vol 10498.34
5: C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV # Mon Aug 1 21:03:32 2022
Spgr 69 - Cell 13.83 25.80 28.73 90.00 90.00 90.00 - Vol 10251.27
6: C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV # Mon Aug 1 21:03:34 2022
Spgr 69 - Cell 14.39 25.16 28.10 90.00 90.00 90.00 - Vol 10173.67
7: C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV # Mon Aug 1 21:03:39 2022
Spgr 69 - Cell 13.54 25.23 27.30 90.00 90.00 90.00 - Vol 9326.07
# dmax dmin ntot nuniq compl i/sig rmeas CC(1/2) ISa B(ov)
---------------------------------------------------------------------------------
1 9.30 0.80 3938 1852 69.0 3.24 20.2 99.4 11.45 8.21 # C:\demo\edtools_demo_data\stagepos_0325\crystal_0000\SMV\CORRECT.LP
- 0.91 0.85 614 290 74.4 0.86 109.5 81.7
2 10.88 0.80 2203 1029 40.4 3.84 11.7 99.9 27.38 9.78 # C:\demo\edtools_demo_data\stagepos_0341\crystal_0000\SMV\CORRECT.LP
- 1.07 0.98 299 135 41.8 1.04 107.2 76.5
3 7.14 0.80 1466 781 26.2 2.61 18.2 97.2 4.73 7.15 # C:\demo\edtools_demo_data\stagepos_0648\crystal_0001\SMV\CORRECT.LP
- 0.84 0.80 142 92 19.9 0.98 62.3 52.7
4 7.24 0.80 1994 1126 38.5 3.27 11.9 99.5 12.91 8.08 # C:\demo\edtools_demo_data\stagepos_0849\crystal_0000\SMV\CORRECT.LP
- 0.98 0.90 322 166 41.2 1.27 70.2 89.0
5 7.08 0.80 1591 808 28.2 2.88 17.1 98.1 6.24 7.63 # C:\demo\edtools_demo_data\stagepos_0905\crystal_0001\SMV\CORRECT.LP
- 0.90 0.85 254 128 30.4 1.17 42.6 95.4
6 5.12 0.80 1669 851 30.2 3.75 16.8 98.0 6.26 5.76 # C:\demo\edtools_demo_data\stagepos_0980\crystal_0000\SMV\CORRECT.LP
- 0.85 0.80 153 109 25.3 1.34 46.1 68.5
7 5.97 0.80 1620 563 21.7 6.15 8.4 99.8 11.79 7.17 # C:\demo\edtools_demo_data\stagepos_1283\crystal_0001\SMV\CORRECT.LP
- 0.85 0.80 187 78 19.1 2.24 45.1 97.9
Wrote 7 cells to file cells.xlsx
Wrote 7 cells to file cells.yaml
Wrote 7 entries to file filelist.txt (completeness > 10.0%, CC(1/2) > 90.0%)
Most likely lattice types:
1 Lattice type `oF` (spgr: 22) was found 7 times (score: 14481)
** the score corresponds to the total number of indexed reflections.
[ ]:
!edtools.find_cell cells.yaml --cluster --metric lcv
[7]:
Image('find_cell_step5.png', embed=True)
[7]:
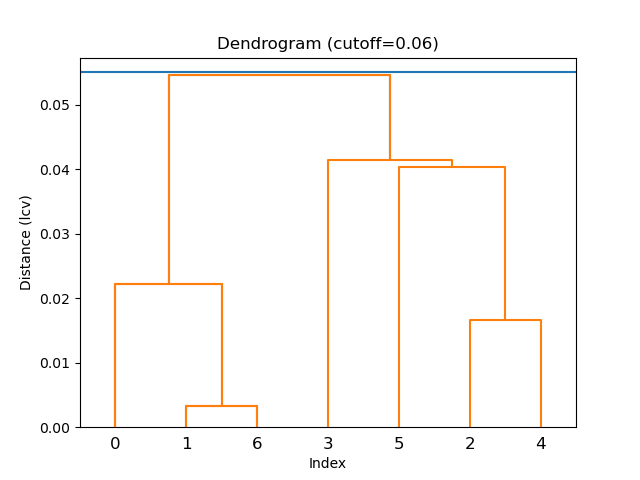
Console Output
Linkage method = average
Cutoff distance = 0.0551
Distance metric = lcv
----------------------------------------
Cluster #1 (7 items)
1 [ 13.97 25.49 27.12 90.00 90.00 90.00] Vol.: 9657.9
2 [ 13.53 25.01 27.18 90.00 90.00 90.00] Vol.: 9195.6
3 [ 14.03 26.02 29.55 90.00 90.00 90.00] Vol.: 10790.3
4 [ 14.94 26.14 26.94 90.00 90.00 90.00] Vol.: 10522.3
5 [ 13.85 25.79 29.03 90.00 90.00 90.00] Vol.: 10364.0
6 [ 14.52 24.95 28.11 90.00 90.00 90.00] Vol.: 10184.6
7 [ 13.53 25.13 27.15 90.00 90.00 90.00] Vol.: 9233.7
---
Mean: [ 14.05 25.50 27.87 90.00 90.00 90.00] Vol.: 9992.6
Min: [ 13.53 24.95 26.94 90.00 90.00 90.00] Vol.: 9195.6
Max: [ 14.94 26.14 29.55 90.00 90.00 90.00] Vol.: 10790.3
Wrote cluster 1 to file `cells_cluster_1_7-items.yaml`
Generate the input file for XSCALE
This command generates the desired unit cell cluster for XSCALE.
[8]:
!edtools.make_xscale cells_cluster_1_7-items.yaml -c 14.05 25.50 27.87 90.00 90.00 90.00 -s 69
Loaded 7 cells
Lowest possible symmetry for 69 (oF): 22
Using:
SPACE_GROUP_NUMBER= 69
UNIT_CELL_CONSTANTS= 14.050 25.500 27.870 90.000 90.000 90.000
Wrote file XSCALE.INP
Wrote file XDSCONV.INP
Run XSCALE
XSCALE calculates the correlation coefficients between different datasets.
[9]:
!wsl xscale
***** XSCALE ***** (VERSION Jan 10, 2022 BUILT=20220220) 1-Aug-2022
Author: Wolfgang Kabsch
Copy licensed until 31-Mar-2023 to
academic users for non-commercial applications
No redistribution.
******************************************************************************
CONTROL CARDS
******************************************************************************
SNRC= 2
SAVE_CORRECTION_IMAGES= FALSE
SPACE_GROUP_NUMBER= 69
UNIT_CELL_CONSTANTS= 14.050 25.500 27.870 90.000 90.000 90.000
OUTPUT_FILE= MERGED.HKL
INPUT_FILE= edtools_demo_data/stagepos_0325/crystal_0000/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_0341/crystal_0000/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_0648/crystal_0001/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_0849/crystal_0000/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_0905/crystal_0001/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_0980/crystal_0000/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
INPUT_FILE= edtools_demo_data/stagepos_1283/crystal_0001/SMV/XDS_ASCII.HKL
INCLUDE_RESOLUTION_RANGE= 20 0.8
THE DATA COLLECTION STATISTICS REPORTED BELOW ASSUMES:
SPACE_GROUP_NUMBER= 69
UNIT_CELL_CONSTANTS= 14.05 25.50 27.87 90.000 90.000 90.000
ALL DATA SETS WILL BE SCALED TO edtools_demo_data/stagepos_0325/crystal_0000/SMV/XDS_ASCII.HKL
******************************************************************************
READING INPUT REFLECTION DATA FILES
******************************************************************************
DATA MEAN REFLECTIONS INPUT FILE NAME
SET# INTENSITY ACCEPTED REJECTED
1 0.3010E+02 3938 0 edtools_demo_data/stagepos_0325/crystal_0000/SMV/XDS_ASCII.HKL
2 0.1368E+02 2205 0 edtools_demo_data/stagepos_0341/crystal_0000/SMV/XDS_ASCII.HKL
3 0.9168E+02 1453 0 edtools_demo_data/stagepos_0648/crystal_0001/SMV/XDS_ASCII.HKL
4 0.4279E+02 1931 0 edtools_demo_data/stagepos_0849/crystal_0000/SMV/XDS_ASCII.HKL
5 0.8542E+02 1590 0 edtools_demo_data/stagepos_0905/crystal_0001/SMV/XDS_ASCII.HKL
6 0.1676E+03 1662 0 edtools_demo_data/stagepos_0980/crystal_0000/SMV/XDS_ASCII.HKL
7 0.1915E+03 1620 0 edtools_demo_data/stagepos_1283/crystal_0001/SMV/XDS_ASCII.HKL
******************************************************************************
OVERALL SCALING AND CRYSTAL DISORDER CORRECTION
******************************************************************************
CORRELATIONS BETWEEN INPUT DATA SETS AFTER CORRECTIONS
DATA SETS NUMBER OF COMMON CORRELATION RATIO OF COMMON B-FACTOR
#i #j REFLECTIONS BETWEEN i,j INTENSITIES (i/j) BETWEEN i,j
1 2 119 0.936 3.1896 0.1394
1 3 87 0.980 0.5337 -0.4854
2 3 164 0.945 0.1982 -0.5975
1 4 216 0.925 1.0428 -0.6421
2 4 116 0.972 0.3076 -0.4582
3 4 131 0.894 1.6716 0.0043
1 5 80 0.959 0.3779 0.1598
2 5 147 0.970 0.1872 -0.3822
3 5 218 0.988 0.9850 0.0336
4 5 96 0.928 0.6114 0.1442
1 6 206 0.955 0.1970 -0.4620
2 6 81 0.949 0.0917 -0.7741
3 6 91 0.934 0.6363 -0.3722
4 6 106 0.927 0.2401 -0.0297
5 6 81 0.866 0.5334 -0.1957
1 7 35 0.965 0.4893 -1.1586
2 7 131 0.981 0.1113 -1.0633
3 7 158 0.984 0.5449 -0.3181
4 7 45 0.833 0.4069 -0.1510
5 7 196 0.987 0.5649 -0.4493
6 7 67 0.846 1.6099 -0.5928
K*EXP(B*SS) = Factor applied to intensities
SS = (2sin(theta)/lambda)^2
K B DATA SET NAME
1.000E+00 0.000 edtools_demo_data/stagepos_0325/crystal_0000/SMV/XDS_ASCII.HKL
2.961E+00 0.170 edtools_demo_data/stagepos_0341/crystal_0000/SMV/XDS_ASCII.HKL
5.374E-01 -0.365 edtools_demo_data/stagepos_0648/crystal_0001/SMV/XDS_ASCII.HKL
9.426E-01 -0.465 edtools_demo_data/stagepos_0849/crystal_0000/SMV/XDS_ASCII.HKL
5.063E-01 -0.243 edtools_demo_data/stagepos_0905/crystal_0001/SMV/XDS_ASCII.HKL
2.304E-01 -0.530 edtools_demo_data/stagepos_0980/crystal_0000/SMV/XDS_ASCII.HKL
3.491E-01 -0.812 edtools_demo_data/stagepos_1283/crystal_0001/SMV/XDS_ASCII.HKL
******************************************************************************
CORRECTION PARAMETERS FOR THE STANDARD ERROR OF REFLECTION INTENSITIES
******************************************************************************
The variance v0(I) of the intensity I obtained from counting statistics is
replaced by v(I)=a*(v0(I)+b*I^2). The model parameters a, b are chosen to
minimize the discrepancies between v(I) and the variance estimated from
sample statistics of symmetry related reflections. This model implicates
an asymptotic limit ISa=1/SQRT(a*b) for the highest I/Sigma(I) that the
experimental setup can produce (Diederichs (2010) Acta Cryst D66, 733-740).
Often the value of ISa is reduced from the initial value ISa0 due to systematic
errors showing up by comparison with other data sets in the scaling procedure.
(ISa=ISa0=-1 if v0 is unknown for a data set.)
a b ISa ISa0 INPUT DATA SET
3.014E+00 1.258E-02 5.14 11.45 edtools_demo_data/stagepos_0325/crystal_0000/SMV/XDS_ASCII.HKL
2.201E+00 3.743E-03 11.02 27.38 edtools_demo_data/stagepos_0341/crystal_0000/SMV/XDS_ASCII.HKL
8.809E+00 2.191E-02 2.28 4.73 edtools_demo_data/stagepos_0648/crystal_0001/SMV/XDS_ASCII.HKL
6.242E+00 1.032E-02 3.94 12.91 edtools_demo_data/stagepos_0849/crystal_0000/SMV/XDS_ASCII.HKL
7.668E+00 1.817E-02 2.68 6.25 edtools_demo_data/stagepos_0905/crystal_0001/SMV/XDS_ASCII.HKL
1.379E+01 1.128E-02 2.53 6.26 edtools_demo_data/stagepos_0980/crystal_0000/SMV/XDS_ASCII.HKL
7.838E-01 1.921E-01 2.58 11.79 edtools_demo_data/stagepos_1283/crystal_0001/SMV/XDS_ASCII.HKL
FACTOR TO PLACE ALL DATA SETS TO AN APPROXIMATE ABSOLUTE SCALE 0.143057E+03
(ASSUMING A PROTEIN WITH 50% SOLVENT)
******************************************************************************
STATISTICS OF SCALED OUTPUT DATA SET : MERGED.HKL
FILE TYPE: XDS_ASCII MERGE=FALSE FRIEDEL'S_LAW=TRUE
13 OUT OF 14399 REFLECTIONS REJECTED
14386 REFLECTIONS ON OUTPUT FILE
******************************************************************************
DEFINITIONS:
R-FACTOR
observed = (SUM(ABS(I(h,i)-I(h))))/(SUM(I(h,i)))
expected = expected R-FACTOR derived from Sigma(I)
COMPARED = number of reflections used for calculating R-FACTOR
I/SIGMA = mean of intensity/Sigma(I) of unique reflections
(after merging symmetry-related observations)
Sigma(I) = standard deviation of reflection intensity I
estimated from sample statistics
R-meas = redundancy independent R-factor (intensities)
Diederichs & Karplus (1997), Nature Struct. Biol. 4, 269-275.
CC(1/2) = percentage of correlation between intensities from
random half-datasets. Correlation significant at
the 0.1% level is marked by an asterisk.
Karplus & Diederichs (2012), Science 336, 1030-33
Anomal = percentage of correlation between random half-sets
Corr of anomalous intensity differences. Correlation
significant at the 0.1% level is marked.
SigAno = mean anomalous difference in units of its estimated
standard deviation (|F(+)-F(-)|/Sigma). F(+), F(-)
are structure factor estimates obtained from the
merged intensity observations in each parity class.
Nano = Number of unique reflections used to calculate
Anomal_Corr & SigAno. At least two observations
for each (+ and -) parity are required.
cpu time used by XSCALE 0.2 sec
elapsed wall-clock time 0.2 sec
Intensity-based clustering
Run intensity-based clustering to further filter out datasets with low correlation (to remove poor quality datasets), or from a different phase that with similar enough unit cell. Cut-off on the dendrogram is selected manually. A number below 0.4 can be a good starting choice.
In the end, integration results from datasets corresponding to different clusters are automatically copied to different folders after running clustering. The merged intensities in file shelx.hkl can be used for structure determination.
[ ]:
!edtools.cluster
[10]:
Image('intensity_cluster.png', embed=True)
[10]:
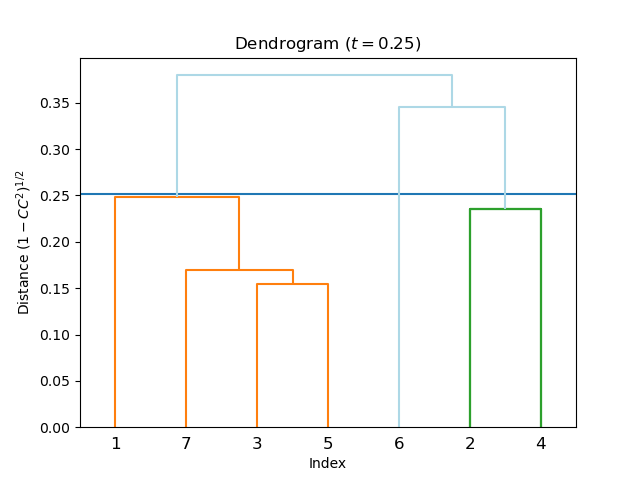
Console Output
Running XSCALE on cluster 1
Running XSCALE on cluster 2
Clustering results
Cutoff distance: 0.252
Equivalent CC(I): 0.968
Method: average
# N_clust CC(1/2) N_obs N_uniq N_poss Compl. N_comp R_meas d_min i/sigma | Lauegr. prob. conf. idx
2** 2 99.8* 4111 1546 2789 55.4 3723 0.143* 0.80 3.27
1*** 4 97.3* 8599 2496 2782 89.7* 8220 0.270* 0.80 2.85
(Sorted by 'Completeness')
Cluster 1: [1, 3, 5, 7]
Cluster 2: [2, 4]
Instruction for using on your own data
Install edtools and all software dependencies on your system
Put all your 3D ED datasets in one folder. All the 3D ED datasets are expected to be in some XDS readable image format, e.g. SMV. A correctly configured XDS.INP file is also expected for each dataset.
Open Windows command prompt from the root directory which contains all the datasets
Follow the demo